In a recent paper in Molecular Ecology on species delimitation, we invoke the age-old criterion of diagnosability to identify lineages that are candidates as name-bearing taxa. For SNP markers, fixed allelic differences between populations or metapopulations provide diagnosability. We apply these markers to a case study of a freshwater turtle, Emydura macquorii, whose systematics has so far defied resolution, to bring to light a dynamic system of substantive allopatric lineages diverging on independent evolutionary trajectories, but held back in the process of speciation by low level and episodic exchange of alleles across drainage divides on various timescales.
In a recent paper in Molecular Ecology on species delimitation, we invoke the age-old criterion of diagnosability to identify lineages that are candidates as name-bearing taxa. For SNP markers, fixed allelic differences between populations or metapopulations provide diagnosability. We apply these markers to a case study of a freshwater turtle, Emydura macquorii, whose systematics has so far defied resolution, to bring to light a dynamic system of substantive allopatric lineages diverging on independent evolutionary trajectories, but held back in the process of speciation by low level and episodic exchange of alleles across drainage divides on various timescales.
Species delimitation is a controversial topic, and most modern approaches use phylogenenetic methods to identify substantive lineages, which are often considered to be species. Jeet Sukumaran and Lacey Knowles (PNAS 2017) argue that dense spatial sampling of individuals and genomes is delivering datasets where the primary structuring is a mix between lineages that have undergone speciation those that reflect spatial structuring of populations.
We elaborate on these concerns. Phylogenetic approaches recover a bifurcating pattern of ancestry and descent and in addition to the concerns of Sukumaran and Knowles, do not accommodate protracted processes of episodic exchange of alleles and resultant partial or complete reticulation in ancestry and descent that we have observed and inferred for in our taxon.
A more rigorous framework is needed to assess the taxonomic status of significant lineages uncovered by increasingly sophisticated molecular data and analyses (see also Singhal et al., 2018). Such a framework will need to incorporate data that considers contemporary, possibly episodic, gene flow as well as lineage divergence and historical patterns of ancestry and descent among populations.
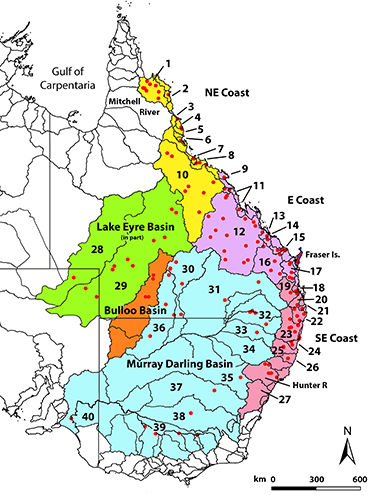
One of the few downsides of the age of molecular phylogenetics is that our ability to generate simple gene and lineage bifurcating phylogenies from ever-increasingly complex genetic and genomic data sets has tended to reduce our focus on one of the most fundamental concepts in species delimitation, that of diagnosability. The identification of diagnostic traits, possessed by all members of a species to the exclusion of all others, has been a cornerstone of species delimitation for centuries -- Linnaeus referred to such traits as the “essential, fixed, intrinsic marks”.
Our strategy requires that unequivocally diagnosable OTUs are identified early in the analysis of species boundaries. As such, it differs from the conventional lineage approach (de Queiroz et al., 1998) in that we place our threshold at the time and place where fixed allelic differences have evolved, rather than immediately at the point of initial lineage divergence, which is always difficult to clearly identify given the often observed gene tree/species tree disparity.
We thus differ in placing greater emphasis on diagnosability (fixed differences) rather than on divergence per se (which draws also from allele frequency differences).
Our strategy is to:
- Achieve comprehensive coverage of populations within the target taxa, both to avoid interpreting sparse sampling of demes on a cline as distinct taxa and to capture evidence of any gene flow across contact zones.
- Identify putative instances of contemporary gene flow between adjacent populations be identified-in our case, qualitatively by examination of ordination plots and quantitatively using software such as NewHybrids
- Identify diagnosable units using analysis of fixed allelic differences, recursively amalgamating populations into operational taxonomic units (diagnosable OTUs).
- Either (a) apply phylogenetic techniques to identify a pattern of ancestry and descent among the OTUs, confident that the process by which they arose was bifrucating, or (b) apply phylogenetic analysis to the sampling sites in the usual way then map the diagnosible OTUs on to the phylogeny to identify diagnosible lineages.
- In a final, and somewhat subjective step, exercise judgement to decide which of the diagnosable OTUs are significant taxonomically (as species, subspecies, ESUs), taking into account the evidence of gene flow between them (or lack of it when opportunity presents, e.g., parapatry), phylogeny and the level of genetic divergences in comparison with other well-defined comparable taxa.
Check out our paper to see how it is done.
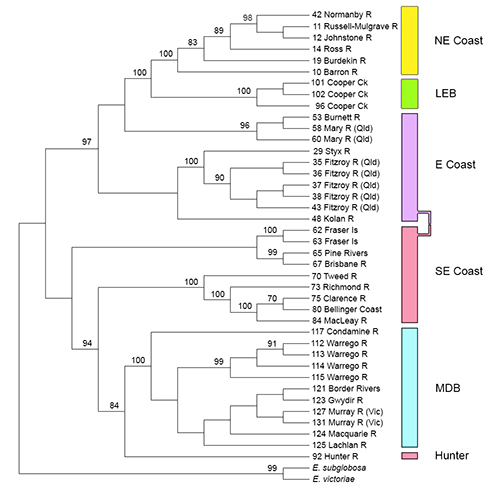
We believe that our strategy and greater focus on diagnosability adds an additional level of objectivity to species delimitation, and makes a potentially important contribution to any rigorous framework for judgements on species boundaries.
References
Georges, A., Gruber, B., Pauly, G.B., White, D., Adams, M., Young, M.J., Kilian, A., Zhang, X., Shaffer, H.B. and Unmack, P. (2018). Genomewide SNP markers breathe new life into phylogeography and species delimitation for the problematic short-necked turtles (Chelidae: Emydura) of eastern Australia. Molecular Ecology, https://onlinelibrary.wiley.com/doi/pdf/10.1111/mec.14925.
Singhal, S., Hoskin, C. J., Couper, P., Potter, S., & Moritz, C. (2018). A framework for resolving cryptic species: A case study from the lizards of the Australian wet tropics. Systematic Biology 67:1061-1075.
Sukumaran, J. and Knowles, L.L. (2017). Multispecies coalescent delimits structure, not species. Proceedings of the National Academy of Sciences 114:1607-1612. https://doi.org/10.1073/pnas.1607921114