A Fresh Look at the Northern Emydura
Understanding the evolutionary history of diversifying lineages and the delineation of species remain major challenges for evolutionary biology.
In a paper we recently submitted, we use single nucleotide polymorphisms (SNPs) and sequence fragment presence-absence (SilicoDArT) data to combine phylogenetics and population genetics to assess species boundaries and challenge current and proposed taxonomies in a genus of Australian freshwater turtles (Chelidae: Emydura) from northern Australia and southern New Guinea.
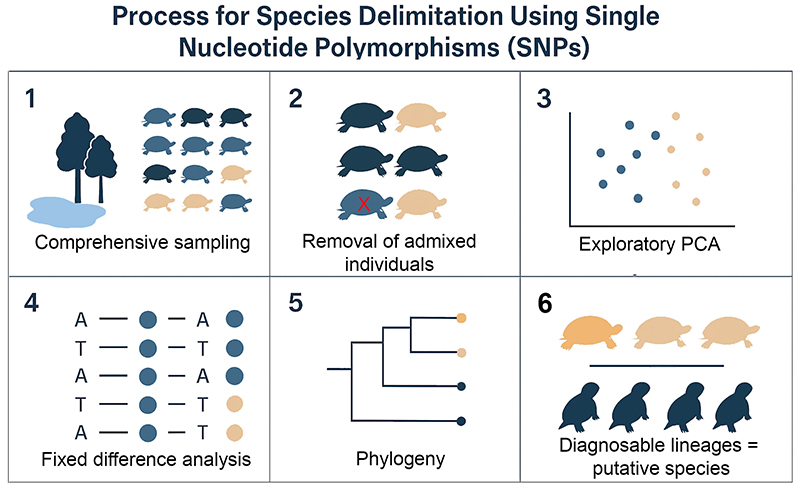
We apply a five-step process – first we identify and remove animals showing evidence of recent or contemporary admixture between putative species. Then we use principal components analysis in an exploratory analysis to identify groupings based on allele frequency structure. An analysis of fixed allelic differences is used to identify diagnosable aggregations of individuals and populations. This analysis assumes no a priori assignment to species. We then identify lineages using phylogenetic analysis. In a final step, we determine which of those lineages are substantive diagnosable lineages. These are the candidate species.
Four currently recognised taxa are identified as substantive diagnosable lineages, two of which we elevate to species status. So we now have four species in the north of Australia and southern New Guinea -- the northern redfaced turtle Emydura australis, the northern yellowfaced turtle Emydura tanybaraga, the diamondhead (so called by tourist operators) Emydura worrelli and the pinted turtle Emydura subglobosa.

Emydura australis [Photo Matt Young, Kimberley] is a senior synonym to Emydura victoriae -- these are not two separate species. The latter name should no longer be used (see also Kehlmaier et al., 2024). Emydura australis occurs in the rivers that drain the Kimberley plateau from the Fitzroy River in the west to the Ord River in the east. It is also found in the Victoria River of the Northern Territory and the Daly River also of the Northern Territory. The Daly River population is enignmatic in that their genotypes have the signature of admixture with Emydura tanybaraga which is likely both historical and contemporary. The diagnostic morphological feature is an expanded mouth plate on the roof of the mouth, presumably an adaptation to moluscivory.

Emydura tanybaraga [Photo Jason Shaffer, Wenlock River] has been poorly defined, based on relatively few specimens. It has also been difficult to distinguish from Emydura worrelli because of the exceptional variability in morphology and coloration of the two species. Both are typically yellow-faced. Here we confirm the disjunct distribution of the species. There are the Northern Territory representatives in the lowland reaches of rivers such as the Finniss-Reynolds, Daly River, South Alligator River and rivers draining Arhnem Land to the north; and there are the Queensland representatives in the rivers draining Cape York to the west into the Gulf of Carpentaria, and to the east in the nortern Cape beyond the distributional range of Emydura macquarii krefftii. There has been an incursion across the range from the Mitchell River into the Barron River (Tinaroo Dam) and evidence of hybridization and introgression with Emydura macquarii krefftii in the Barron and Russell-Mulgrave Rivers.

Emydura worrelli [Photo CCNT, McArthur River] is elevated to a species in its own right based on our evidence, but following the earlier judgements of Cann and Sadlier (2018). It occupies principally the rivers flowing north and west into the Gulf of Carpentaria, from the Roper River in the west to the Flinders River in the East. It also occurs in the Daly River above the escarpment (e.g. Sleisbeck) and is restricted in this way in other rivers draining Arhnem Land west of the Roper River. There is clear evidence of admixture between E. worrelli and E. tanybaraga where they co-occur, notably in the Flinders River.

Emydura subglobosa [Photo Arthur Georges, Fly River] has its stronghold in southern New Guinea in the Rivers flowing south from but not including the Langurru Belt in the west to the Vanapa region in the east. Its range extends into Australia at the tip of Cape York in the Jardine River. It is interesting because our nuclear markers show that it clearly belongs with the New Guinea populations, presumably a recent incursion to Australia during the last glacial cycle. The interesting thing about this is that the Jardine River population appear to have captured the Emydura tanybaraga mitochondrial haplotype. So the mitochondiral phylogeny and the SNP and SilicoDArT nuclear DNA phylogeny recover very different phylogenetic signals. We have seen this before with Chelodina expansa and C. longicollis and C. canni (Hodges, 2015).
Is the Jardine River population a subspecies? It depends on your definition of subspecies. My view is that subspecies tend to be conveniences to assist with communication -- they do not have to be clades (that is, monophyletic), can be defined on a simple morphological character (e.g. yellow eyestripe in adults), and the criterion of diagnosability is relaxed. Their value is in grouping populations on the basis of morphological similarities to the exclusion of other populations. There is little value in assigning a subspecies name to an entity that is found only in one place -- you may as well just refer to the Australian population of Emydura subglobosa. There are differing views on this.

The only area of real contention is in regard to the Emydura in the Mitchell River of Western Australia. In our study, we had four individuals we caught in the plunge pool of Little Mitchell Falls. They were adult but diminutive. Three had red eyestripes and one had a yellow eyestripe. All had the diagnostic expanded mouth plate diagnostic of Emydura australis. These were odd in several respects. They were clearly embedded in the E. australis clade in the nuclear phylogeny but marginally diagnosable from Emydura australis (p=0.0495 < 0.05) as an internal subclade. Their mitochondrial haplotypes were more closely aligned with Emydura tanybaraga than E. australis. Consistent with this, they represented the only population in the study we examined that showed evidence of outbreeding.
When we combine our results with those of Kehlmaier et al. (2024), we conclude that there are two taxa with highly divergent mitochondrial haplotypes that are subject to admixture in the Mitchell River of the Kimberley. One is clearly Emydura australis, and we assign the other as depicted to Emydura tanybaraga [Photo Jiri Lochman, Mitchell River, WA]. Emydura tanybaraga thus has a disjunct distribution across northern Australia with one population in the Mitchell River, a group of populations in the lowland reaches of rivers in the Northern Territory, and another group of populations on Cape York of Queensland. The Mitchell River situation requires further attention.
We note that the lineage structure revealed by analysis of nuclear markers differed in important respects from a mitochondrial phylogeny. We attribute this to recent or contemporary lateral transfer of mitochondria during hybridization events, deeper historical hybridization or possibly incomplete lineage sorting of the mitochondrial genome.
Taxonomic decisions in cases of allopatry require subjective judgement. Our five-step strategy adds an additional level of objectivity before that subjectivity is applied, and so reduces the risk of taxonomic inflation that can accompany lineage approaches to species delimitation.
Hybridization and associated introgression presents particular challenges for those working with morphological data. The ability to detect hybrid individuals could be of enormous benefit in future morphological studies, permitting the detection and removal of individuals whose morphological attributes are a consequence of their hybrid origin. Undetected hybridization and introgression can confound morphological discrimination between species and render difficult the formal diagnoses and description of species. We believe that the framework of relationships we present is an excellent platform for assessing and describing the extent of intraspecific morphological variation within taxa well grounded in molecular evidence, and vis-à-vis, how best to describe a species and the extent of interspecific morphology between it and other taxa. As Ross Sadlier put it to me, there has never been a better time to commence such morphological studies given the current resolution provided by genetic studies, the collections now available to work on around Australia, and the availability of computing software to analyse the morphological data and ask questions of it.
Note: This account is based on an article available as yet only as a preprint, not yet having benefitted from rigorous peer review. As such, the contents of the article have not yet contributed to the scientific record. It may be subject to change. Indeed, if you have any constructive suggestions, pass them on.
- Arthur Georges, Peter J. Unmack, Andrzej Kilian, Xiuwen Zhang, Yolarnie Amepou and Duminda S.B. Dissanayake. 2025.Lineages as species or lineages within species – using diagnosability to better inform species delimitation (Chelidae: Emydura). Preprint.