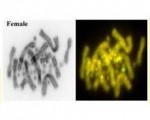
Who would have thought you could pull DNA from historical museum specimens dating back to 1794 and sequence it? Well it can be done to generate special insights to the confusing world of taxonomy, clarifying type specimens and relationships among species. Here a team from the Museum of Zoology of Senckenberg Dresden has joined forces with a team from the Institute of Applied Ecology, University of Canberra, to bring together whole mitochondrial sequences from living populatons of Australasian Chelid turtle with those generated from museum type specimens. We gang up with some other taxonomists to solve some longstanding taxonomic enigmas.
Results of our study appeared this week in Scientific Reports.
For readers interested in the general implications and not on the specifics, we make the case that a central challenge for taxonomy has arisen from the rapid advances in DNA technologies.These technologies have revolutionized taxonomy by generating a large suite of new characters to complement those available from more traditonal morphological analysis. The challenge is to match these new insights from molecular biology with previous taxonomic decisions based on name-bearing type specimens.
There are a very many taxonomic issues arising from, by modern standards, poor historical species descriptions. Unfortunately, museum type specimens are either fixed in formalin, a process that cross-links the DNA with associated proteins and renders the DNA inaccessible, or they are preserved in such a way that DNA is highly degraded.
Although still not routine, generating historical DNA data has become feasible on a large scale. DNA sequencing of name-bearing type specimens is thus coming to be regarded as the gold standard for taxonomy. It enables unambiguous assignment of extant populations to the named entity or enables a clear distinction between newly discovered forms and those that have already been described.
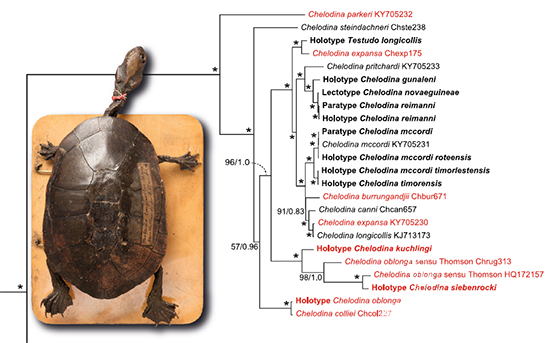
It is in this context that we generated whole mitochondrial sequences from historical type specimens of a range of Australasian chelid freshwater turtles, and addressed a number of long-standing taxonomic issues.
Perhaps the most controversial issue to be resolved was that of the names applied to the snake-necked turtles of Northern Australia and the south west of Western Australia. Without boring you with the details, a morphological analysis found that the holotype of Chelodina oblonga -- the Oblong Turtle well known in the Perth region -- was most likely a specimen of the northern species, Chelodina rugosa. This was supported by evidence drawn from the travels of the person who collected the original specimen.

For the nomenclaturalist, this meant that the name Chelodina oblonga should be used for the snake-necked turtle in northern Australia, setting aside the established name of Chelodina rugosa, and that a new name, Chelodina colliei, the next with precedence, should be assigned to the snakeneck from the Perth region. A petition was made to the International Commission on Zoological Nomenclature (ICZN) to keep the status quo, but the petition failed. Many were upset with the change.
What does the mitogenome of the type specimen tell us?
The holotype of Chelodina oblonga has a mitochondrion that is clearly from the south-western lineage, and so the decision to assign the type to the northern lineage, and sink Chelodina rugosa, is not supported by our data. So as not to further destablize the taxonomy, we retain Chelodina colliei for the southwestern lineage and resurrect Chelodina rugosa for the northern lineage. The name Chelodina oblonga, for reasons we outline, is sent into obscurity as a nomen dubium.
Hopefully, this puts to bed a controversy that has lasted 130 years.
Another controversial issue surrounds Chelodina kuchlingi. This species has been regarded variously as a valid taxon or not, because it was described from a single specimen of uncertain provenance, without a scientifically defensible diagnosis or description. Our data support C. kuchlingi as a valid species. However, the stated type locality of Kalumbaru is in doubt, and we were unable to link the taxon to any population of Chelodina. Locating populations of this species is thus a high priority.

For the short-necked species, we were able to confirm the assignment of the Myuchelys species from the Gwydir and Namoi Rivers of the Murray-Darling Basin to the holotype of Phrynops bellii previously of unknown provenance, as suggested by John Cann after some meticulous sleuthing on the travels of the person who collected the holotype.
The northern Emydura are a complete mess, with historical types from the southern Murray-Darling basin thrown inadvertently into the mix and some unhelpful contributions made outside the mainstream peer-reviewed literature. We are at last able to sort this out, at least partially.
The name Emydura victoriae is unambiguously assigned to the red-faced Emydura species from the Victoria River, Northern Territory. Emydura australis of uncertain geographic provenance is considered to be a valid species. As a result, there remains the complication presented by Emydura australis, which has precedence over Emydura victoriae should the two be found to be one taxon. The two taxa are very close.
There is evidence of historical mitochondrial introgression and several other results of taxonomic significance that you can find in our paper which is open access.
Happy reading!