Methods used in routine identification of bacterial pathogens in medical microbiology can include a combination of biochemical tests, mass spectrometry, barcode sequencing (e.g. 16S rRNA) or whole genome sequencing. In a paper that came out today in the Journal of Microbiological Methods, CONACYT PhD student Berenice Talamantes explores the utility of a new approach -- reduced representation sequencing. She has also applied the technique to environmental microbes in work yet to appear.
Extensive publicly-available databases of DNA sequences from pathogenic bacteria have been amassed in recent years. This provides an opportunity for using bacterial genome sequencing for identification purposes. Whole genome sequencing is increasing in popularity, although at present it remains a relatively expensive approach to bacterial identification and typing. DNA barcoding has been automated, but has its own particular challenges.
An alternative is to obtain a representative sample of the bacterial genome and use that for species and strain identification. The representation is obtained using what are called restriction enzymes. These enzymes are found in bacteria and provide a defence mechanism against invading viruses. They cut up into fragments the alien viral DNA that enters the bacterial cell. To identify the virus these enzymes cut at or near specific recognition sites in the DNA code of the virus. This makes them immensely useful.
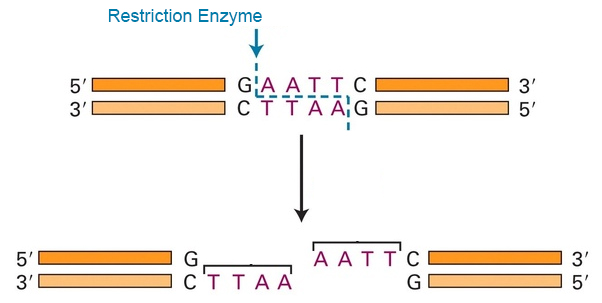 Scientists have put these 'cutters' to use in subsampling genomes that are too big or unwieldy to sequence in their entirity. Companies have become established to provide high-throughput representational sequencing for agriculture, animal breeding, biodiversity assessment and wildlife forensics. Once such company, Diversity Arrays Technology, based at the University of Canberra, provides a service called DArTseq to generate reproducable representations of genomes of a wide variety of organisms.
Scientists have put these 'cutters' to use in subsampling genomes that are too big or unwieldy to sequence in their entirity. Companies have become established to provide high-throughput representational sequencing for agriculture, animal breeding, biodiversity assessment and wildlife forensics. Once such company, Diversity Arrays Technology, based at the University of Canberra, provides a service called DArTseq to generate reproducable representations of genomes of a wide variety of organisms.
Berenice worked with Diversity Arrays Technology to apply representational sequencing to the challenging problem of identifying microbial species and strains in two contexts -- identifying bacterial pathogens in a hospital, and identifying thermophilic bacteria in the environment.
There are thousands of restriction enzymes known in bacteria, and over 600 have been turned to use as commercially available restriction enzymes. Berenice chose pairs of enzymes -- PstI with MseI, PstI with HpaII and MseI with HpaII -- in a double digest of the target genomes. Double digestion produces thousands of specific fragments of DNA from the target bacteria in a very reproducible manner. These fragments, if they are of a manageable size, can be sequenced and compared to public databases for identification to species.
Alignment of the DArTseq sequences to the known sequences in the public database were processed using a bioinformatics pipeline developed by Berenice and her colleague Jason Carling -- Currito3.1 DNA Fragment Analysis Software. It automates what is a rather complex bioinformatic challenge of matching the DArTseq sequences to those of known targets.
All 165 isolates sourced a public hospital in Canberra were correctly identified to genus and species by each of the three combinations of restriction enzymes. Berenice also had considerable success in identifying bacteria to strain.
The high level of agreement between bacterial identification using reduced-representation genome sequencing and standard hospital identification indicates that this new approach has the potential as an alternative to established techniques and more exhaustive approaches like whole genome sequencing. The key limitation is the quality of the reference database, particularly in regards to strain typing. Development and access to a comprehensive and taxonomically accurate bacterial genome sequence database containing relevant bacterial species and strains will be a key ingredient in the successful adoption of this approach.
You can read all about Berenice's work in the latest issue of the Journal of Microbiological Methods, 160:11-19. Feel free to contact Berenice by email for further information. She is currently in Mexico.