Mention species concept nowadays and people are likely throw up their hands and give a negative or dismissive reaction. This is because it has been such a contentious issue and because almost every argument that could be mounted in support of one idea over another has already been discussed ad nausium. Most people are simply tired of the endless arguments.
Mention species concept nowadays and people are likely throw up their hands and give a negative or dismissive reaction. This is because it has been such a contentious issue and because almost every argument that could be mounted in support of one idea over another has already been discussed ad nausium. Most people are simply tired of the endless arguments.
Nevertheless we have another go in our recent paper which appeared this week in the Biological Journal of Linnean Society.
There appear to be two fundamentally divergent views. One is that species emerge from the various mechanisms that lead to reproductive isolation. Once such species are established, they are lineages on independent evolutionary trajectories regardless of any geographical happenstance that may bring two descendent species back into contact. So well-defined species are necessarily reproductively isolated. Speciation is a process not an event, and involves the evolution of traits in each incipeient species that retard gene flow and introgression between the two should they come into contact. Extinction is termination of the lineage when the last individual perishes.
The difficulty with this "Biological Species Concept" is in its application. While genetic structure in the context of long-standing microsympatry is solid evidence of species delimitation, there are major issues with applying the concept in cases of allopatry. How much genetic divergence is required to infer that two lineages have achieved reproductive incompatability -- clearly that requires subjective judgement to make the operational decision. After all, the two -- reproductive isolation and divergence across neutral loci -- are not necessarily closely linked. Furthermore, speciation is a process -- how far down the process must two divergent lineages pass before they are regarded as distinct species. Again, subjective judgement required. Under this concept, all species are lineages and not all lineages are species, but how do you call one over the other.
 The second view of species dispenses with the criterion of reproductive isolation as necessary, and regards all substantive lineages on independent evolutionary trajectories as species. This concept, operationally at least, includes as species lineages that are on independent evolutionary trajectories by virtue of geographic isolation and lineages on independent evolutionary trajectories by virtue of reproductive isolation. Speciation is more closely aligned with an event, that is cladogenesis, rather than a protracted process, and extinction can occur when two lineages come again into contact and homogenize.
The second view of species dispenses with the criterion of reproductive isolation as necessary, and regards all substantive lineages on independent evolutionary trajectories as species. This concept, operationally at least, includes as species lineages that are on independent evolutionary trajectories by virtue of geographic isolation and lineages on independent evolutionary trajectories by virtue of reproductive isolation. Speciation is more closely aligned with an event, that is cladogenesis, rather than a protracted process, and extinction can occur when two lineages come again into contact and homogenize.
Although the lineage species concept is operationally more satisfactory, by sidestepping the difficulty in demonstrating reproductive isolation, it presents a challenge of how to distinguish divergence of lineages that are species with divergence of lineages within species (see Sukumaran and Knowles, 2017). Trying to interpret this statement will highlight the clash of mindsets.
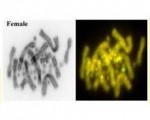
While not pretending to have solved this conundrum, we argue that the concept of diagnosability should be central to any criterion for delimiting species. That is, every individual of a species should be able to be assigned to that species by one or more diagnostic characters. It is necessary but not sufficient that a lineage be diagnosable from all other lineages if it is to be regarded as a distinct species. Some such lineages and those that are not characterised by diagnostic characters represent variation within species. In the context of SNP datasets (or for that matter the earlier allozyme datasets), a fixed allelic difference at a locus is a diagnostic character. Assuming it is not an artifact of limited sampling, a fixed allelic difference allows one to unambiguously assign an individual to its species.
We thus present a framework to interrogate genomic datasets and coalescent-based species trees for the presence of candidate species using genetic data alone. We propose six steps:
- visualize genetic affinities among individuals using PCA to identify both discrete groups and individuals involved in contemporary admixture to be held aside for independent consideration (see Marshall et al., 2021);
- apply phylogenetic techniques to identify lineages;
- use fixed allelic differences to assess diagnosability of those lineages as potential candidate species (see also Georges et al. 2018);
- interpret the diagnosable lineages in a geographical context (sympatry, parapatry, allopatry);
- assess significance of difference or trends in the context of sampling intensity (see Chambers and Hillis, 2020); and
- adopt a holistic approach to available evidence to inform decisions on species status in the difficult cases of allopatry.
We adopt this approach to distinguish candidate species from within-species lineages for a widespread species complex of Australian freshwater fishes (genus Retropinna).
Our framework addresses two cornerstone issues in systematics that are often not discussed explicitly in genomic species discovery: diagnosability and how to determine it, and what criteria should be used to decide whether diagnosable lineages are conspecific or represent different species.