Innumerable approaches to analyse genetic data are now available to guide conservation, ecological and agricultural projects. However, streamlined and accessible tools are needed to bring these approaches within reach of a broader user base. dartR was released in 2018 to lessen the intrinsic complexity of analysing single nucleotide polymorphisms (SNPs) and dominant markers (presence/absence of amplified sequence tags) by providing user-friendly data quality control and marker selection functions. dartR users have grown steadily since its release and provided valuable feedback on their interaction with the package allowing us to enhance dartR capabilities.
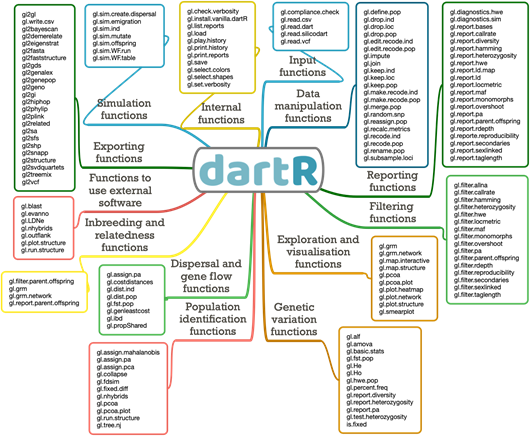 We are pleased to announce the release of dartR version 2. We have substantially increased the number of functions from 45 to 144 to enhance the user experience by extending plot customisation, function standardisation, increasing user support and function speed. dartR now provides many additional functions for importing data, exporting data and linking to other packages -- an easy-to-navigate conduit between data generation and analysis options already available via other packages. We also implemented simulation functions whose results can be analysed seamlessly with several other dartR functions.
We are pleased to announce the release of dartR version 2. We have substantially increased the number of functions from 45 to 144 to enhance the user experience by extending plot customisation, function standardisation, increasing user support and function speed. dartR now provides many additional functions for importing data, exporting data and linking to other packages -- an easy-to-navigate conduit between data generation and analysis options already available via other packages. We also implemented simulation functions whose results can be analysed seamlessly with several other dartR functions.
Version 2 is on CRAN and can be accessed using the instructions on the dartR web page. An updated series of tutorials is also available.
As more methods and approaches mature to inform conservation, we envision that accessible platforms to analyse genetic data will play a crucial role in translating science into practice. We look forward to hearing more feedback from users through the dartR google group.

The Core dartR team. [30-Mar-22]